The immune system was originally regarded as a host defense mechanism for recognizing and eliminating invading pathogens. As our knowledge of the immune system continues to grow, it has become increasingly appreciated that the immune system plays a much broader role in regulating tissue maintenance and coordinating tissue function. Efficient function of the immune system requires a delicate balance between activation and suppression of various cell types, pathways, and mediators while avoiding detrimental reactions to self and innocuous antigens. Deviation from such balance underlies autoimmune and inflammatory diseases, infection-associated immunopathology, cancer, transplant rejection, cardiovascular diseases, and neurodegenerative disorders.
Our lab studies how immune responses are regulated by regulatory (Treg) T cells, a dominant cell population suppressing a wide range of immune effectors. Treg cells have also been implicated in tissue repair and regeneration through direct interactions with non-immune cells. Currently, we are focusing on understanding universal and tissue-specific mechanisms that regulate Treg biology, which could help in conceiving better strategies for targeting Treg cells for therapeutic purposes. Some of the ongoing projects in the lab focus on elucidating the biochemical, molecular, and cellular basis of immune regulation by studying common and context-dependent regulations of Treg cell gene expression and function.
1. How do Treg cells maintain their identity and tune their function in the context of ongoing inflammation?
Precise control of the magnitude and duration of an immune response is vital for optimizing pathogen clearance while minimizing immunopathologies to the host. Understanding the cross-talk between immune activation and Treg-mediated suppression will be highly informative for designing adequate therapies for autoimmunity, infection associated cytokine release syndrome, and cancer. Our previous work has demonstrated that a single cohort of Treg cells can function in established autoimmune inflammation, causing its reversal, and persist stably to provide long-term protection. In the future, we hope to obtain a more generalizable understanding of how Treg cells maintain their identity, functionality, and long-term survival in the context of inflammation. Specifically, we want to answer questions such as: What type of inflammation preserves/destabilizes Treg cells? What cell type/soluble factors in the inflammatory environment enhance/attenuate Treg cell lineage stability and functionality? How are Treg cells maintained during repetitive immunological challenges in the periphery when thymic output is limited?
2. In non-lymphoid tissues, how do immune cells adapt to the local microenvironment to control tissue inflammation and regulate tissue homeostasis?
Following initial priming, diverse immune cell types, including Treg cells, migrate to non-lymphoid tissues where they contribute to tissue-resident immunity and assist tissue function. Tissue-resident immune cells must adapt metabolically to stay fit and execute their function in response to changes in nutrient and oxygen availability in the tissue microenvironment. We are curious to understand how changes in metabolism influence cellular physiology at the chromatin, transcript, and protein level. We found that protein O-GlcNAcylation, a post-translational modification (PTM) regulated by inputs from multiple metabolic pathways, is selectively upregulated in tissue-resident immune cells and may link metabolic reprogramming to changes in gene transcription. We ask: How does this metabolically sensitive PTM impact the function of Treg cells in infection, autoimmunity, and cancer? How does O-GlcNAcylation on key transcription factors program immune cells during tissue adaptation?
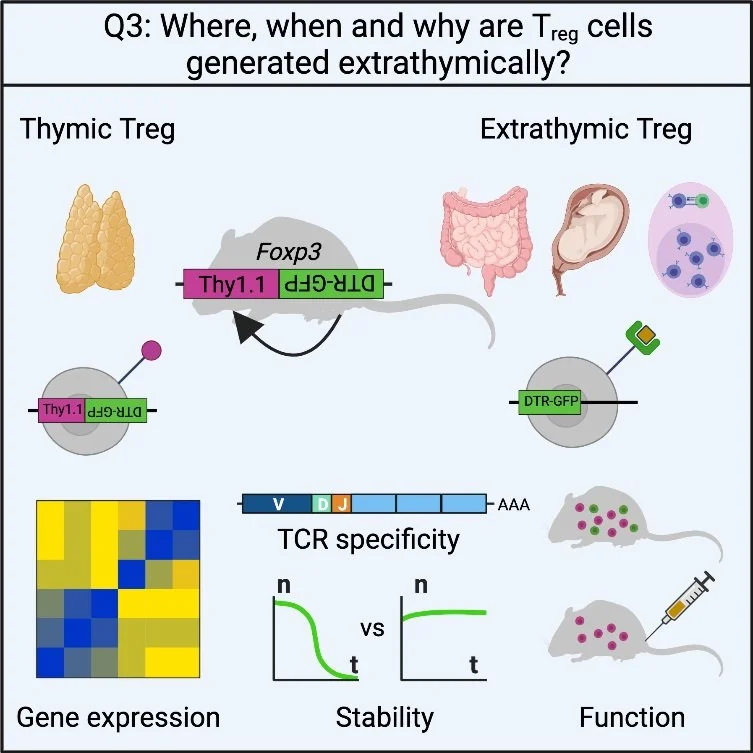
3. Where, when, and why are Treg cells generated extrathymically?
The majority of Treg cells develop in the thymus (tTreg), which is indispensable for preventing fatal autoimmunity. However, Treg cells that develop extrathymically (pTreg) are much less well understood except for those in the intestine. Based on some latest findings, we hypothesize that pTreg cells in other organs and contexts, even relatively minor in numerical terms, could play a significant role in immune regulation. By implementing novel genetic tools, we want to explore: Where and when are pTreg generated in the periphery? What are the properties and functions of pTreg cells in these contexts? Are there human counterparts of these phenomena?
4. Method development
We are always enthusiastic about developing and utilizing new tools and methods to further immunology research. We are eager to adopt newly developed molecular biology methods, particularly low input and single cell technologies, to decode gene regulation in T cells. We are also experienced in creating new mouse models with complex genetic circuits, many of which allow for spatial and temporal manipulation of cells, proteins, and mRNA. We are on the way to generating better tools to answer some of the outstanding questions in T cell biology, including but not limited to: What is the role of TCR specificity in Treg cell mediated immune regulation? What is the origin, longevity, and migration pattern of tumor-infiltrating T cells? What is the contribution of T cells and other immune cells to maternal-fetal tolerance? Are there novel immune regulators among previously uncharacterized biomolecules?